Several times a year I am asked about a small segment match. For the purposes of this blog, I am considering anything below 8 centimorgans (cM) a small segment.
Many experienced and eloquent genetic genealogists have written on this topic. The titles of their articles include a spoiler alert:
- Leah Larkin Low Matches Lie
- Leah Larkin The Small Segment Debate Is Over
- Blaine Bettinger An In-depth Analysis of the Use of Small Segments as Genealogical Evidence
- Roberta Estes Identical by Descent, State, Population and Chance
Measuring DNA
To understand why small segments are deceptive, it’s important to understand how DNA matching is measured. When you look at a DNA match list at any company, you will see the shared cM amount between you and your match. But that’s not all they use to determine if you are a match. Each company has a threshold for sharing which incorporates shared cMs and other characteristics of the DNA data. This may include matching on one or both chromosomes (remember you get one from each parent), the SNP density (referring to how rich the genetic information is – a very complex topic I don’t pretend to understand), the number of segments you share, and endogamy. Endogamy is the situation where groups of people, often geographically or culturally isolated, partnered with each other over hundreds or thousands of years. The result is that you can have many relationships with your matches, which inflates the amount of shared DNA. If you come from an endogamous population, you need to share more DNA in larger segments to be considered a DNA match. The ISOGG wiki has a table that compares the DNA testing companies matching criteria in detail. Search for “Criteria for matching segments” in the ISOGG Wiki link.
In genetic genealogy, we create images of shared cMs that make measuring DNA look like a simple thing you could do with a ruler, with coloured bars indicating shared segments of DNA. In the image below, I’ve used DNA Painter, importing data from known paternal DNA matches in the blue shaded portion near the top of the image and known maternal matches in the pink section in the lower part of the image. Each different coloured bar represents a different shared ancestor.

The most basic definition of shared cM dances around the issue of what is actually being measured:
cM: “a unit of measure for autosomal DNA segments. The more DNA we share with someone in centimorgans, the more closely related we are.” Leah Larkin PhD
These simple images and definitions cover up the wealth of science behind what is actually being measured. The ISOGG Wiki provides the following definition for centimorgan:
A centimorgan…is a unit of recombinant frequency which is used to measure genetic distance. It is often used to imply distance along a chromosome, and takes into account how often recombination occurs in a region. A region with few cMs undergoes relatively less recombination. The number of base pairs to which it corresponds varies widely across the genome (different regions of a chromosome have different propensities towards crossover). One centiMorgan corresponds to about 1 million base pairs in humans on average. The centiMorgan is equal to a 1% chance that a marker at one genetic locus on a chromosome will be separated from a marker at a second locus due to crossing over in a single generation.
Whoa! This definition reminds me that I will never understand everything there is about genetic genealogy, and that there are scientists who developed the strategies the DNA testing companies use. Blaine Bettinger has summarized company information on segments and matching here. I have to keep reminding myself that cM measurement isn’t like the length of ribbon, it’s about how likely it is that the DNA will split apart when an egg or sperm is being made.
False matching
Even with all of this science, a match on your match list at a DNA testing company might not be real. How can that be?
That small segment might be a pseudosegment, a false segment which leads to false matches. This can happen because the DNA company takes your DNA apart into the two chromosomes and puts it back together again. Sometimes it is put back together wrong, weaving back and forth between the father’s and mother’s DNA. The smaller the segment, the greater the chance it is a pseudosegment. (For more information and an illustration, see Identical By Descent in the ISOGG Wiki.)
If you have transferred your DNA to another testing company (FamilyTreeDNA and MyHeritage allow DNA transfers) or you are using the third-party site, GEDmatch, your small segment might be due to imputation or be from a known pile-up region.
Imputation. Whenever anyone does a DNA test, there are some regions that can’t be read (“no-calls”) and those sections are estimated (imputed). This happens at every DNA testing company. If a DNA company has changed their testing chip over the years they use imputation to allow them to analyze the DNA in the sections of the chip that differ. For companies that accept transfers and GEDmatch, there will always be imputation because of the number of different chips they are analyzing and comparing to each other. Every company does imputation using the methods their scientists have developed. Imputation can create a small segment and it can also separate a larger segment into two segments. Roberta Estes has a three part series in her blog on imputation starting here.
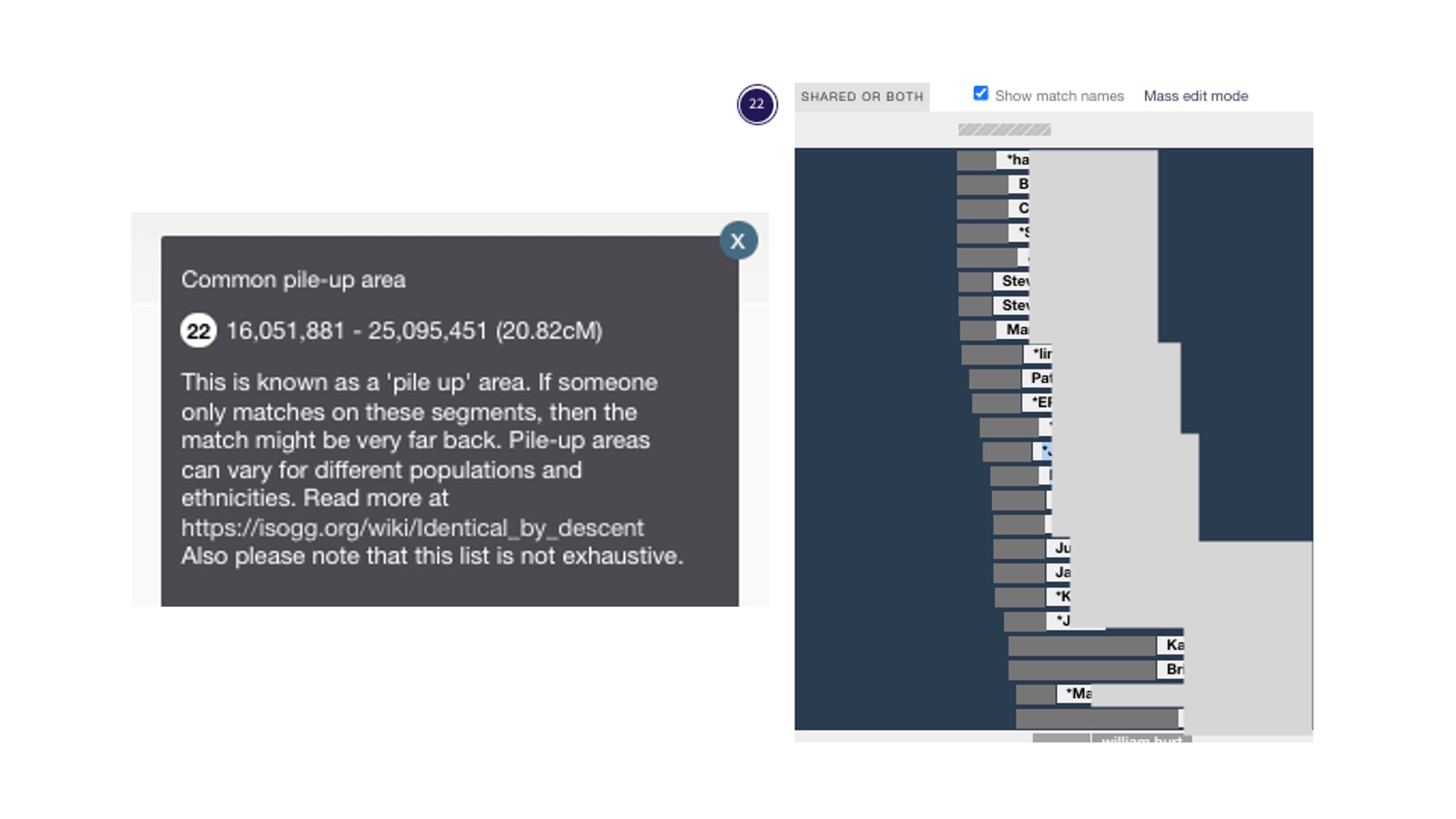
There are pile-up regions where many people have the same DNA. These are also called excess IBD (Identical by Descent). Testing companies don’t report those regions (they have algorithms that leave them out of DNA matching), but GEDmatch reports these regions. More on that in the ISOGG Wiki. Jonny Perl has included the known pile-up areas in DNA Painter; a grey bar with horizontal stripes appears above each chromosome where they are known to happen. In the image above of chromosome 1, you can see the grey bar in the middle above the blue-shaded area. When you click on the grey bar, you will find additional information about that pile-up region. I’ve provided a close-up below of a pile-up region for Chromosome 22. The explanatory text box is on the left. The painting on the right shows data from GEDmatch. In 2023, I shared DNA in this pile-up region with 75 people. I know it’s a pile-up area because there are so many matches and I can see the grey bar with the diagonal stripes at the top. I don’t consider these DNA matches even though they share 10-14 cM with me on that chromosome.
Two additional complications with small segments
Many people do DNA testing to learn more about their country or region origin. Some companies call this ethnicity results. The best term for this kind of data is biogeographical ancestry – where your distant ancestors were at a point in time. That information is also in our chromosomes. That’s complication number one. Your small segment could be Identical by Population, as described by Roberta Estes. Everyone or almost everyone who descends from people on the same migratory population path for thousands of years has the same segment of DNA.
Complication number two pertains to your goal. Many of us do DNA testing because we want to find our ancestors and give them names. We can do that if our matches occur within a genealogical time frame, defined as the time when their might be documents to help us. Your small segment could be from an ancestor not within a genealogical time frame. Using simulated DNA data, Leah Larkin has found a 10 cm match could be a 9th cousin, meaning you share 8x great-grandparents. For most of us, this is at the edge of documentary genealogy. A smaller segment, such as 7 cM could be from a 10th great-grandparent or a 40th great-grandparent or there is a 58% chance it is false. (Simulated data from Leah Larkin. Data on false segment size is from Tim Jantzen in the ISOGG Wiki. See Blaine Bettinger for company specific information on false segment sizes. )
At the third party DNA site, GEDmatch, you can alter the matching thresholds to below what the testing companies are doing. This is where the danger lies. Just because you can set a lower threshold doesn’t mean you should.
But I match someone with a LOT of small segments!
If all of the segments are small, the most likely explanation is endogamy. As mentioned earlier, the strategy for working with endogamous communities is to use larger segments and avoid the small ones. If you are working with an endogamous community, you will be applying different strategies to analyze your DNA. Paul Woodbury has a two part series Dealing with Endogamy. He also lectures and teaches courses. You can seek out presentations and courses by Dr. Adina Newman. Diahan Southard offers an Endogamy Course (full disclosure, I work for Diahan Southard as a coach). Leah Larkin, the DNA Geek, periodically offers an Endogamy lecture and writes about Endogamy in her blog. I recommend all of these from personal experience.
A rational approach to using segment data
With documentary genealogy, we know we need to start with the present and work our way back. You can do the same thing in genetic genealogy using segment data. Jim Bartlett, author of the blog segment-ology, calls this “walking the segment back.”
Let me introduce you to some of my ancestors and DNA-tested cousins in the image below. Skip to the next paragraph if this type of family tree diagram is familiar to you. If it’s not, what follows is a description of the diagram and a reminder of relationship terminology and abbreviations. In the image, I’m at the bottom in a light blue box. My dad is immediately above me, then my granddad, then my great-grandparents, Walter Hale Davis and May (Hilton) Davis in green. All the cousins that I share with the ancestral couple of Walter and May are in green. SG, CP, MR, and I are second cousins, because second cousins share great-grandparents. PK, CP’s parent, my dad, MR’s parent, and JS are all first cousins to each other because they share Walter and Mary as grandparents. Since I am one generation younger than these first cousins, I am their first cousin once removed (1C1R). Moving up the diagram, Walter’s parents were William Hale Davis and Sarah Jane (Ellis) Davis. A descendant of Walter’s sibling has also done a DNA test. RD is shown in a light green to match William and Sarah Jane. Since William and Sarah Jane are RD’s great-grandparents and RD is one generation older than me, we are second cousins once removed (2C1R). And up at the top are my 3x-great grandparents, Rev. T.O. Ellis, MD and his wife, Elizabeth (Long) Ellis, in the dark green. I share this ancestral couple (T.O. and Elizabeth) with two cousins (siblings, GS and DM) also in dark green in the lower right. T.O. and Elizabeth are the 2x-great grandparents of GS and DM, so we are third cousins once removed (3C1R).
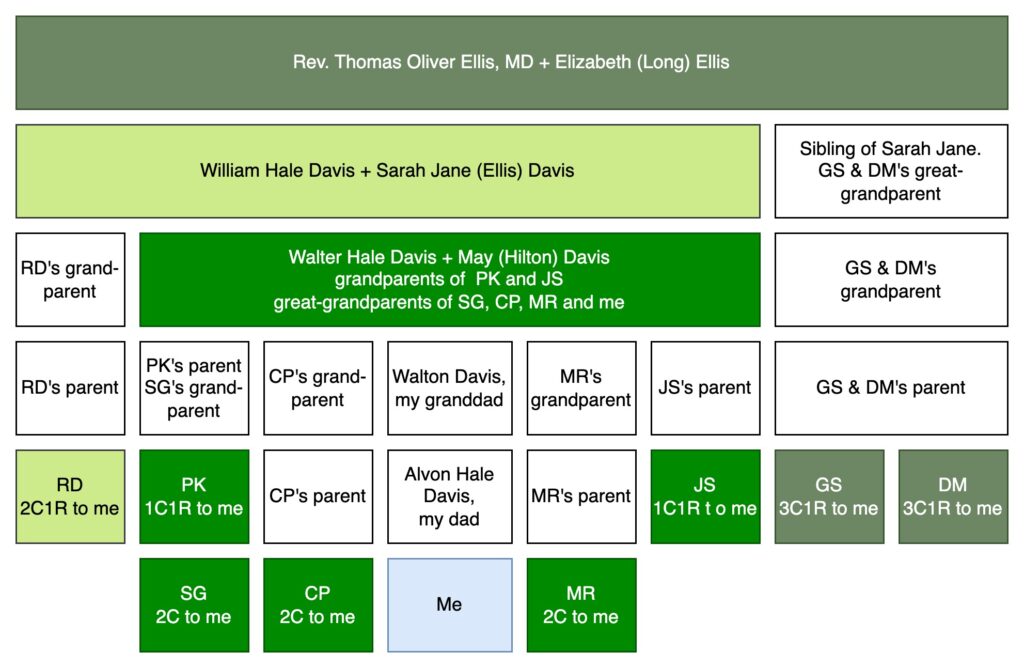
MyHeritage and FamilyTreeDNA allow DNA testers to download the segments you share with your DNA matches. If you know your relationship to a DNA match, you can assign the segment to an ancestral couple. In the image below of Chromosome 1, I started by “painting” the DNA from my great-grandparents, Walter Hale Davis and May (Hilton) Davis with green. You always receive DNA from great-grandparents, so that’s a great place to start painting your DNA. I painted the segment data from two 1C1R (PK and JS) and three 2C (SG, CP and MR). I didn’t really need SG since their parent has also tested, but it is a good illustration of how DNA segments tend to get smaller every generation. The lightest green match (RD) is a 2C1R who shares my 2x great-grandparents, William Hale Davis and Sarah Jane (Ellis) Davis. If you look at the comparison of PK to RD, you can see that RD is contained within the green segment from my great-grandparents. This makes sense. The DNA from PK came from either my Davis ancestor or my Hilton ancestor, and it’s clear that most or all of it came from Davis, since I don’t share Hilton ancestors with RD. Then I have two siblings, DM (13.2 cM) and GS (11.8 cM) who both descend from my 3x-great grandparents, Rev. Thomas Oliver Ellis, MD and Elizabeth (Long) Ellis. The same pattern holds: the segment fits within the segment from RD, who is both a Davis and an Ellis. Dark green is either Ellis or Long or both.

I may some day find a cousin who descends from Rev. Ellis’ father, Josiah Shelton Ellis, or more distant ancestors, but the chances get increasingly remote as we go further back in time. If an ancestor has no or few siblings, the line could have died out. If there are recent immigrants, they may not be in the testing databases. By using this methodology, I can be more confident that a smaller segment came from a more distant ancestor. Note: The smallest segments I painted are both over 10 cM and came from 3x great-grandparents and 3C1R matches. The average segment size for a 3C1R is 16 cM based on simulated data from Leah Larkin.
If you are interested in using segment data, consider encouraging your matches to upload to MyHeritage. Why? It’s free to upload your DNA, they have good privacy protections, and in addition to being able to gather the data for chromosome painting, there are other useful tools for genealogy at MyHeritage.
The constant plea: Test the oldest generations of your family now!
We can enhance our reach by testing the oldest generation. They will have larger segments to work with and are one step closer to your ancestors. If you have any older relatives (parents, aunts, uncles, cousins one generation older), buy a DNA test for them. (Watch for sales!) Then visit them personally and make sure they do all the steps for the DNA kit to be activated and usable.
Summary
Genetic genealogists avoid using small segments when making genealogical conclusions. There is science behind the limitations of DNA matching. Genetic genealogy needs to be treated like documentary research: start with the present and work your way back.
